- Protein domain
-
A protein domain is a part of protein sequence and structure that can evolve, function, and exist independently of the rest of the protein chain. Each domain forms a compact three-dimensional structure and often can be independently stable and folded. Many proteins consist of several structural domains. One domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. Domains vary in length from between about 25 amino acids up to 500 amino acids in length. The shortest domains such as zinc fingers are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins.
Contents
Background
The concept of the domain was first proposed in 1973 by Wetlaufer after X-ray crystallographic studies of hen lysozyme [1] and papain [2] and by limited proteolysis studies of immunoglobulins.[3][4] Wetlaufer defined domains as stable units of protein structure that could fold autonomously. In the past domains have been described as units of:
Each definition is valid and will often overlap, i.e. a compact structural domain that is found amongst diverse proteins is likely to fold independently within its structural environment. Nature often brings several domains together to form multidomain and multifunctional proteins with a vast number of possibilities.[8] In a multidomain protein, each domain may fulfill its own function independently, or in a concerted manner with its neighbours. Domains can either serve as modules for building up large assemblies such as virus particles or muscle fibres, or can provide specific catalytic or binding sites as found in enzymes or regulatory proteins.
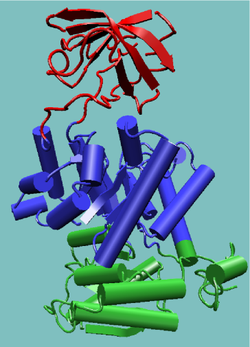
An appropriate example is pyruvate kinase, a glycolytic enzyme that plays an important role in regulating the flux from fructose-1,6-biphosphate to pyruvate. It contains an all-β regulatory domain, an α/β-substrate binding domain and an α/β-nucleotide binding domain, connected by several polypeptide linkers [9] (see figure, right). Each domain in this protein occurs in diverse sets of protein families.
The central α/β-barrel substrate binding domain is one of the most common enzyme folds. It is seen in many different enzyme families catalysing completely unrelated reactions.[10] The α/β-barrel is commonly called the TIM barrel named after triose phosphate isomerase, which was the first such structure to be solved.[11] It is currently classified into 26 homologous families in the CATH domain database.[12] The TIM barrel is formed from a sequence of β-α-β motifs closed by the first and last strand hydrogen bonding together, forming an eight stranded barrel. There is debate about the evolutionary origin of this domain. One study has suggested that a single ancestral enzyme could have diverged into several families,[13] while another suggests that a stable TIM-barrel structure has evolved through convergent evolution.[14]
The TIM-barrel in pyruvate kinase is 'discontinuous', meaning that more than one segment of the polypeptide is required to form the domain. This is likely to be the result of the insertion of one domain into another during the protein's evolution. It has been shown from known structures that about a quarter of structural domains are discontinuous.[15][16] The inserted β-barrel regulatory domain is 'continuous', made up of a single stretch of polypeptide.
Covalent association of two domains represents a functional and structural advantage since there is an increase in stability when compared with the same structures non-covalently associated.[17] Other, advantages are the protection of intermediates within inter-domain enzymatic clefts that may otherwise be unstable in aqueous environments, and a fixed stoichiometric ratio of the enzymatic activity necessary for a sequential set of reactions.[18]
Domains are units of protein structure
Main article: Protein structureThe primary structure (string of amino acids) of a protein ultimately encodes its uniquely folded 3D conformation.[19] The most important factor governing the folding of a protein into 3D structure is the distribution of polar and non-polar side chains.[20] Folding is driven by the burial of hydrophobic side chains into the interior of the molecule so to avoid contact with the aqueous environment. Generally proteins have a core of hydrophobic residues surrounded by a shell of hydrophilic residues. Since the peptide bonds themselves are polar they are neutralised by hydrogen bonding with each other when in the hydrophobic environment. This gives rise to regions of the polypeptide that form regular 3D structural patterns called secondary structure. There are two main types of secondary structure: α-helices and β-sheets.
Some simple combinations of secondary structure elements have been found to frequently occur in protein structure and are referred to as supersecondary structure or motifs. For example, the β-hairpin motif consists of two adjacent antiparallel β-strands joined by a small loop. It is present in most antiparallel β structures both as an isolated ribbon and as part of more complex β-sheets. Another common super-secondary structure is the β-α-β motif, which is frequently used to connect two parallel β-strands. The central α-helix connects the C-termini of the first strand to the N-termini of the second strand, packing its side chains against the β-sheet and therefore shielding the hydrophobic residues of the β-strands from the surface.
Structural alignment is an important tool for determining domains.
Tertiary structure of domains
Several motifs pack together to form compact, local, semi-independent units called domains.[5] The overall 3D structure of the polypeptide chain is referred to as the protein's tertiary structure. Domains are the fundamental units of tertiary structure, each domain containing an individual hydrophobic core built from secondary structural units connected by loop regions. The packing of the polypeptide is usually much tighter in the interior than the exterior of the domain producing a solid-like core and a fluid-like surface.[21] In fact, core residues are often conserved in a protein family, whereas the residues in loops are less conserved, unless they are involved in the protein's function. Protein tertiary structure can be divided into four main classes based on the secondary structural content of the domain.[22]
- All-α domains have a domain core built exclusively from α-helices. This class is dominated by small folds, many of which form a simple bundle with helices running up and down.
- All-β domains have a core composed of antiparallel β-sheets, usually two sheets packed against each other. Various patterns can be identified in the arrangement of the strands, often giving rise to the identification of recurring motifs, for example the Greek key motif.[23]
- α+β domains are a mixture of all-α and all-β motifs. Classification of proteins into this class is difficult because of overlaps to the other three classes and therefore is not used in the CATH domain database.[12]
- α/β domains are made from a combination of β-α-β motifs that predominantly form a parallel β-sheet surrounded by amphipathic α-helices. The secondary structures are arranged in layers or barrels.
Domains have limits on size
Domains have limits on size.[24] The size of individual structural domains varies from 36 residues in E-selectin to 692 residues in lipoxygenase-1,[15] but the majority, 90%, have less than 200 residues[25] with an average of approximately 100 residues.[26] Very short domains, less than 40 residues, are often stabilised by metal ions or disulfide bonds. Larger domains, greater than 300 residues, are likely to consist of multiple hydrophobic cores.[27]
Domains and quaternary structure
Many proteins have a quaternary structure, which consists of several polypeptide chains that associate into an oligomeric molecule. Each polypeptide chain in such a protein is called a subunit. Hemoglobin, for example, consists of two α and two β subunits. Each of the four chains has an all-α globin fold with a heme pocket.
Domain swapping is a mechanism for forming oligomeric assemblies.[28] In domain swapping, a secondary or tertiary element of a monomeric protein is replaced by the same element of another protein. Domain swapping can range from secondary structure elements to whole structural domains. It also represents a model of evolution for functional adaptation by oligomerisation, e.g. oligomeric enzymes that have their active site at subunit interfaces.[29]
Domains as evolutionary modules
Nature is a tinkerer and not an inventor,[30] new sequences are adapted from pre-existing sequences rather than invented. Domains are the common material used by nature to generate new sequences, they can be thought of as genetically mobile units, referred to as 'modules'. Often, the C and N termini of domains are close together in space, allowing them to easily be "slotted into" parent structures during the process of evolution. Many domain families are found in all three forms of life, Archaea, Bacteria and Eukarya. Domains that are repeatedly found in diverse proteins are often referred to as modules, examples can be found among extracellular proteins associated with clotting, fibrinolysis, complement, the extracellular matrix, cell surface adhesion molecules and cytokine receptors.[31]
Molecular evolution gives rise to families of related proteins with similar sequence and structure. However, sequence similarities can be extremely low between proteins that share the same structure. Protein structures may be similar because proteins have diverged from a common ancestor. Alternatively, some folds may be more favored than others as they represent stable arrangements of secondary structures and some proteins may converge towards these folds over the course of evolution . There are currently about 45,000 experimentally determined protein 3D structures deposited within the Protein Data Bank (PDB).[32] However this set contains a lot of identical or very similar structures. All proteins should be classified to structural families to understand their evolutionary relationships. Structural comparisons are best achieved at the domain level. For this reason many algorithms have been developed to automatically assign domains in proteins with known 3D structure, see 'Domain definition from structural co-ordinates'.
The CATH domain database classifies domains into approximately 800 fold families, ten of these folds are highly populated and are referred to as 'super-folds'. Super-folds are defined as folds for which there are at least three structures without significant sequence similarity.[33] The most populated is the α/β-barrel super-fold as described previously.
Multidomain proteins
The majority of genomic proteins, two-thirds in unicellular organisms and more than 80% in metazoa, are multidomain proteins created as a result of gene duplication events.[34] Many domains in multidomain structures could have once existed as independent proteins. More and more domains in eukaryotic multidomain proteins can be found as independent proteins in prokaryotes.[35] For example, vertebrates have a multi-enzyme polypeptide containing the GAR synthetase, AIR synthetase and GAR transformylase modules (GARs-AIRs-GARt; GAR: glycinamide ribonucleotide synthetase/transferase; AIR: aminoimidazole ribonucleotide synthetase). In insects, the polypeptide appears as GARs-(AIRs)2-GARt, in yeast GARs-AIRs is encoded separately from GARt, and in bacteria each domain is encoded separately.[36]
Origin
Multidomain proteins are likely to have emerged from a selective pressure during evolution to create new functions. Various proteins have diverged from common ancestors by different combinations and associations of domains. Modular units frequently move about, within and between biological systems through mechanisms of genetic shuffling:
- transposition of mobile elements including horizontal transfers (between species);[37]
- gross rearrangements such as inversions, translocations, deletions and duplications;
- homologous recombination;
- slippage of DNA polymerase during replication.
Types of organisation
The simplest multidomain organisation seen in proteins is that of a single domain repeated in tandem.[38] The domains may interact with each other or remain isolated, like beads on string. The giant 30,000 residue muscle protein titin comprises about 120 fibronectin-III-type and Ig-type domains.[39] In the serine proteases, a gene duplication event has led to the formation of a two β-barrel domain enzyme.[40] The repeats have diverged so widely that there is no obvious sequence similarity between them. The active site is located at a cleft between the two β-barrel domains, in which functionally important residues are contributed from each domain. Genetically engineered mutants of the chymotrypsin serine protease were shown to have some proteinase activity even though their active site residues were abolished and it has therefore been postulated that the duplication event enhanced the enzyme's activity.[40]
Modules frequently display different connectivity relationships, as illustrated by the kinesins and ABC transporters. The kinesin motor domain can be at either end of a polypeptide chain that includes a coiled-coil region and a cargo domain.[41] ABC transporters are built with up to four domains consisting of two unrelated modules, ATP-binding cassette and an integral membrane module, arranged in various combinations.
Not only do domains recombine, but there are many examples of a domain having been inserted into another. Sequence or structural similarities to other domains demonstrate that homologues of inserted and parent domains can exist independently. An example is that of the 'fingers' inserted into the 'palm' domain within the polymerases of the Pol I family.[42] Since a domain can be inserted into another, there should always be at least one continuous domain in a multidomain protein. This is the main difference between definitions of structural domains and evolutionary/functional domains. An evolutionary domain will be limited to one or two connections between domains, whereas structural domains can have unlimited connections, within a given criterion of the existence of a common core. Several structural domains could be assigned to an evolutionary domain.
Domains are autonomous folding units
Folding
Further information: Protein foldingProtein folding - the unsolved problem : Since the seminal work of Anfinsen over forty years ago,[19] the goal to completely understand the mechanism by which a polypeptide rapidly folds into its stable native conformation remains elusive. Many experimental folding studies have contributed much to our understanding, but the principles that govern protein folding are still based on those discovered in the very first studies of folding. Anfinsen showed that the native state of a protein is thermodynamically stable, the conformation being at a global minimum of its free energy.
Folding is a directed search of conformational space allowing the protein to fold on a biologically feasible time scale. The Levinthal paradox states that if an averaged sized protein would sample all possible conformations before finding the one with the lowest energy, the whole process would take billions of years.[43] Proteins typically fold within 0.1 and 1000 seconds, therefore the protein folding process must be directed some way through a specific folding pathway. The forces that direct this search are likely to be a combination of local and global influences whose effects are felt at various stages of the reaction.[44]
Advances in experimental and theoretical studies have shown that folding can be viewed in terms of energy landscapes,[45][46] where folding kinetics is considered as a progressive organisation of an ensemble of partially folded structures through which a protein passes on its way to the folded structure. This has been described in terms of a folding funnel, in which an unfolded protein has a large number of conformational states available and there are fewer states available to the folded protein. A funnel implies that for protein folding there is a decrease in energy and loss of entropy with increasing tertiary structure formation. The local roughness of the funnel reflects kinetic traps, corresponding to the accumulation of misfolded intermediates. A folding chain progresses toward lower intra-chain free-energies by increasing its compactness. The chains conformational options become increasingly narrowed ultimately toward one native structure.
Advantage of domains in protein folding
The organisation of large proteins by structural domains represents an advantage for protein folding, with each domain being able to individually fold, accelerating the folding process and reducing a potentially large combination of residue interactions. Furthermore, given the observed random distribution of hydrophobic residues in proteins,[47] domain formation appears to be the optimal solution for a large protein to bury its hydrophobic residues while keeping the hydrophilic residues at the surface.[48][49]
However, the role of inter-domain interactions in protein folding and in energetics of stabilisation of the native structure, probably differs for each protein. In T4 lysozyme, the influence of one domain on the other is so strong that the entire molecule is resistant to proteolytic cleavage. In this case, folding is a sequential process where the C-terminal domain is required to fold independently in an early step, and the other domain requires the presence of the folded C-terminal domain for folding and stabilisation.[50]
It has been found that the folding of an isolated domain can take place at the same rate or sometimes faster than that of the integrated domain.[51] Suggesting that unfavourable interactions with the rest of the protein can occur during folding. Several arguments suggest that the slowest step in the folding of large proteins is the pairing of the folded domains.[27] This is either because the domains are not folded entirely correctly or because the small adjustments required for their interaction are energetically unfavourable,[52] such as the removal of water from the domain interface.
Domains and protein flexibility
The presence of multiple domains in proteins gives rise to a great deal of flexibility and mobility, leading to protein domain dynamics[53]. Domain motions can be inferred by comparing different structures of a protein (as in Database of Molecular Motions), or they can be directly observed using spectra[54][55] measured by neutron spin echo spectroscopy. They can also be suggested by sampling in extensive molecular dynamics trajectories.[56] Domain motions are important for:[57]
- catalysis[citation needed]
- regulatory activity
- transport of metabolites
- formation of protein assemblies
- cellular locomotion
One of the largest observed domain motions is the `swivelling' mechanism in pyruvate phosphate dikinase. The phosphoinositide domain swivels between two states in order to bring a phosphate group from the active site of the nucleotide binding domain to that of the phosphoenolpyruvate/pyruvate domain.[58] The phosphate group is moved over a distance of 45A involving a domain motion of about 100 degrees around a single residue.In enzymes, the closure of one domain onto another captures a substrate by an induced fit, allowing the reaction to take place in a controlled way. A detailed analysis by Gerstein led to the classification of two basic types of domain motion; hinge and shear.[57] Only a relatively small portion of the chain, namely the inter-domain linker and side chains undergo significant conformational changes upon domain rearrangement.[59]
Hinges by secondary structures
A study by Hayward[60] found that the termini of α-helices and β-sheets form hinges in a large number of cases. Many hinges were found to involve two secondary structure elements acting like hinges of a door, allowing an opening and closing motion to occur. This can arise when two neighbouring strands within a β-sheet situated in one domain, diverge apart as they join the other domain. The two resulting termini then form the bending regions between the two domains. α-helices that preserve their hydrogen bonding network when bent are found to behave as mechanical hinges, storing `elastic energy' that drives the closure of domains for rapid capture of a substrate.[60]
Helical to extended conformation
The interconversion of helical and extended conformations at the site of a domain boundary is not uncommon. In calmodulin, torsion angles change for five residues in the middle of a domain linking α-helix. The helix is split into two, almost perpendicular, smaller helices separated by four residues of an extended strand.[61][62]
Shear motions
Shear motions involve a small sliding movement of domain interfaces, controlled by the amino acid side chains within the interface. Proteins displaying shear motions often have a layered architecture: stacking of secondary structures. The interdomain linker has merely the role of keeping the domains in close proximity.
Domain motion and functional dynamics in enzymes
The analysis of the internal dynamics of structurally different, but functionally similar enzymes has highlighted a common relationship between the positioning of the active site and the two principal protein sub-domains. In fact, for several members of the hydrolase superfamily, the catalytic site is located close to the interface separating the two principal quasi-rigid domains.[56] Such positioning appears instrumental for maintaining the precise geometry of the active site, while allowing for an appreciable functionally-oriented modulation of the flanking regions resulting from the relative motion of the two sub-domains.
Domain definition from structural co-ordinates
The importance of domains as structural building blocks and elements of evolution has brought about many automated methods for their identification and classification in proteins of known structure. Automatic procedures for reliable domain assignment is essential for the generation of the domain databases, especially as the number of protein structures is increasing. Although the boundaries of a domain can be determined by visual inspection, construction of an automated method is not straightforward. Problems occur when faced with domains that are discontinuous or highly associated.[63] The fact that there is no standard definition of what a domain really is has meant that domain assignments have varied enormously, with each researcher using a unique set of criteria.[64]
A structural domain is a compact, globular sub-structure with more interactions within it than with the rest of the protein.[59] Therefore, a structural domain can be determined by two visual characteristics; its compactness and its extent of isolation.[65] Measures of local compactness in proteins have been used in many of the early methods of domain assignment[66][67][68][69] and in several of the more recent methods.[25][70][71][72][73]
Methods
One of the first algorithms[66] used a Cα-Cα distance map together with a hierarchical clustering routine that considered proteins as several small segments, 10 residues in length. The initial segments were clustered one after another based on inter-segment distances; segments with the shortest distances were clustered and considered as single segments thereafter. The stepwise clustering finally included the full protein. Go[69] also exploited the fact that inter-domain distances are normally larger than intra-domain distances; all possible Cα-Cα distances were represented as diagonal plots in which there were distinct patterns for helices, extended strands and combinations of secondary structures.
The method by Sowdhamini and Blundell clusters secondary structures in a protein based on their Cα-Cα distances and identifies domains from the pattern in their dendrograms.[63] As the procedure does not consider the protein as a continuous chain of amino acids there are no problems in treating discontinuous domains. Specific nodes in these dendrograms are identified as tertiary structural clusters of the protein, these include both super-secondary structures and domains. The DOMAK algorithm is used to create the 3Dee domain database.[71] It calculates a 'split value' from the number of each type of contact when the protein is divided arbitrarily into two parts. This split value is large when the two parts of the structure are distinct.
The method of Wodak and Janin[74] was based on the calculated interface areas between two chain segments repeatedly cleaved at various residue positions. Interface areas were calculated by comparing surface areas of the cleaved segments with that of the native structure. Potential domain boundaries can be identified at a site where the interface area was at a minimum. Other methods have used measures of solvent accessibility to calculate compactness.[25][75][76]
The PUU algorithm[16] incorporates a harmonic model used to approximate inter-domain dynamics. The underlying physical concept is that many rigid interactions will occur within each domain and loose interactions will occur between domains. This algorithm is used to define domains in the FSSP domain database.[70]
Swindells (1995) developed a method, DETECTIVE, for identification of domains in protein structures based on the idea that domains have a hydrophobic interior. Deficiencies were found to occur when hydrophobic cores from different domains continue through the interface region.
RigidFinder is a novel method for identification of protein rigid blocks (domains and loops) from two different conformations. Rigid blocks are defined as blocks where all inter residue distances are conserved across conformations.
A general method to identify dynamical domains, that is protein regions that behave approximately as rigid units in the course of structural fluctuations, has been introduced by Potestio et al.[56] and, among other applications was also used to compare the consistency of the dynamics-based domain subdivisions with standard structure-based ones. The method, termed PiSQRD, is publicly available in the form of a webserver.[77] The latter allows users to optimally subdivide single-chain or multimeric proteins into quasi-rigid domains[56][77] based on the collective modes of fluctuation of the system. By default the latter are calculated through an elastic network model;[78] alternatively pre-calculated essential dynamical spaces can be uploaded by the user.
Example domains
- Armadillo repeats : named after the β-catenin-like Armadillo protein of the fruit fly Drosophila.
- Basic Leucine zipper domain (bZIP domain) : is found in many DNA-binding eukaryotic proteins. One part of the domain contains a region that mediates sequence-specific DNA-binding properties and the Leucine zipper that is required for the dimerization of two DNA-binding regions. The DNA-binding region comprises a number of basic aminoacids such as arginine and lysine
- Cadherin repeats : Cadherins function as Ca2+-dependent cell-cell adhesion proteins. Cadherin domains are extracellular regions which mediate cell-to-cell homophilic binding between cadherins on the surface of adjacent cells.
- Death effector domain (DED) : allows protein-protein binding by homotypic interactions (DED-DED). Caspase proteases trigger apoptosis via proteolytic cascades. Pro-Caspase-8 and pro-caspase-9 bind to specific adaptor molecules via DED domains and this leads to autoactivation of caspases.
- EF hand : a helix-turn-helix structural motif found in each structural domain of the signaling protein calmodulin and in the muscle protein troponin-C.
- Immunoglobulin-like domains : are found in proteins of the immunoglobulin superfamily (IgSF).[79] They contain about 70-110 amino acids and are classified into different categories (IgV, IgC1, IgC2 and IgI) according to their size and function. They possess a characteristic fold in which two beta sheets form a "sandwich" that is stabilized by interactions between conserved cysteines and other charged amino acids. They are important for protein-to-protein interactions in processes of cell adhesion, cell activation, and molecular recognition. These domains are commonly found in molecules with roles in the immune system.
- Phosphotyrosine-binding domain (PTB) : PTB domains usually bind to phosphorylated tyrosine residues. They are often found in signal transduction proteins. PTB-domain binding specificity is determined by residues to the amino-terminal side of the phosphotyrosine. Examples: the PTB domains of both SHC and IRS-1 bind to a NPXpY sequence. PTB-containing proteins such as SHC and IRS-1 are important for insulin responses of human cells.
- Pleckstrin homology domain (PH) : PH domains bind phosphoinositides with high affinity. Specificity for PtdIns(3)P, PtdIns(4)P, PtdIns(3,4)P2, PtdIns(4,5)P2, and PtdIns(3,4,5)P3 have all been observed. Given the fact that phosphoinositides are sequestered to various cell membranes (due to their long lipophilic tail) the PH domains usually causes recruitment of the protein in question to a membrane where the protein can exert a certain function in cell signalling, cytoskeletal reorganization or membrane trafficking.
- Src homology 2 domain (SH2) : SH2 domains are often found in signal transduction proteins. SH2 domains confer binding to phosphorylated tyrosine (pTyr). Named after the phosphotyrosine binding domain of the src viral oncogene, which is itself a tyrosine kinase. See also: SH3 domain.
- Zinc finger DNA binding domain (ZnF_GATA) : ZnF_GATA domain-containing proteins are typically transcription factors that usually bind to the DNA sequence [AT]GATA[AG] of promoters.
See also
- Binding domain
- Short linear motif
- Protein
- Protein structure
- Protein structure prediction
- Protein structure prediction software
- Protein family
- Structural biology
- Structural Classification of Proteins (SCOP)
- CATH
References
This article incorporates text and figures from George, R. A. (2002) "Predicting Structural Domains in Proteins" Thesis, University College London, which were contributed by its author.
- ^ Phillips DC. (1966). "The three-dimensional structure of an enzyme molecule". Scientific American 215 (5): 78–90. PMID 5978599.
- ^ Drenth J, Jansonius JN, Koekoek R, Swen HM, Wolthers BG. (1968). "Structure of papain". Nature 218 (5145): 929–32. doi:10.1038/218929a0. PMID 5681232.
- ^ Porter RR. (1973). "Structural studies of immunoglobulins". Science 180 (4087): 713–6. doi:10.1126/science.180.4087.713. PMID 4122075. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=4122075.
- ^ Edelman GM. (1973). "Antibody structure and molecular immunology". Science 180 (4088): 830–40. doi:10.1126/science.180.4088.830. PMID 4540988.
- ^ a b Richardson J. S. (1981). "The anatomy and taxonomy of protein structure". Adv Protein Chem 34: 167–339. doi:10.1016/S0065-3233(08)60520-3. PMID 7020376. http://kinemage.biochem.duke.edu/teaching/anatax/index.html.
- ^ Bork P. (1991). "Shuffled domains in extracellular proteins". FEBS Lett 286 (1-2): 47–54. doi:10.1016/0014-5793(91)80937-X. PMID 1864378.
- ^ Wetlaufer DB. (1973). "Nucleation, rapid folding, and globular intrachain regions in proteins". Proc Natl Acad Sci USA 70 (3): 697–701. doi:10.1073/pnas.70.3.697. PMC 433338. PMID 4351801. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=433338.
- ^ Chothia C. (1992). "Proteins. One thousand families for the molecular biologist". Nature 357 (6379): 543–4. doi:10.1038/357543a0. PMID 1608464.
- ^ George RA, Heringa J. (2002). "An analysis of protein domain linkers: their classification and role in protein folding". Protein Eng 15 (11): 871–9. doi:10.1093/protein/15.11.871. PMID 12538906.
- ^ Hegyi H, and Gerstein M. (1999). "The relationship between protein structure and function: a comprehensive survey with application to the yeast genome". J Mol Biol 288 (1): 147–64. doi:10.1006/jmbi.1999.2661. PMID 10329133.
- ^ Banner et al.; Bloomer, AC; Petsko, GA; Phillips, DC; Pogson, CI; Wilson, IA; Corran, PH; Furth, AJ et al. (1975). "Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5 angstrom resolution using amino acid sequence data". Nature 255 (5510): 609–614. doi:10.1038/255609a0. PMID 1134550.
- ^ a b Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. (1997). "CATH--a hierarchic classification of protein domain structures". Structure 5 (8): 1093–108. doi:10.1016/S0969-2126(97)00260-8. PMID 9309224.
- ^ Copley, R. R. and Bork, P (2000). "Homology among (betaalpha)(8) barrels: implications for the evolution of metabolic pathways". J Mol Biol 303 (4): 627–641. doi:10.1006/jmbi.2000.4152. PMID 11054297.
- ^ Lesk AM, Brändén CI, Chothia C. (1989). "Structural principles of alpha/beta barrel proteins: the packing of the interior of the sheet". Proteins 5 (2): 139–48. doi:10.1002/prot.340050208. PMID 2664768.
- ^ a b Jones S, Stewart M, Michie A, Swindells MB, Orengo C, Thornton JM. (1998). "Domain assignment for protein structures using a consensus approach: characterization and analysis". Protein Sci 7 (2): 233–42. doi:10.1002/pro.5560070202. PMC 2143930. PMID 9521098. http://www.proteinscience.org/cgi/pmidlookup?view=long&pmid=9521098.
- ^ a b Holm L, Sander C. (1994). "Parser for protein folding units". Proteins 19 (3): 256–68. doi:10.1002/prot.340190309. PMID 7937738.
- ^ Ghélis C, Yon JM. (1979). "[Conformational coupling between structural units. A decisive step in the functional structure formation]". C R Seances Acad Sci D 289 (2): 197–9. PMID 117925.
- ^ Ostermeier M, Benkovic SJ. (2000). "Evolution of protein function by domain swapping". Adv Protein Chem 55: 29–77. PMID 11050932.
- ^ a b ANFINSEN CB, HABER E, SELA M, WHITE FH Jr. (1961). "The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain". Proc Natl Acad Sci USA 47 (9): 1309–14. doi:10.1073/pnas.47.9.1309. PMC 223141. PMID 13683522. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=223141.
- ^ Cordes, M. H., Davidson, A. R., and Sauer, R. T (1996). "Sequence space, folding and protein design". Curr Opin Struct Biol 6 (1): 3–10. doi:10.1016/S0959-440X(96)80088-1. PMID 8696970.
- ^ Zhou, Y., Vitkup, D., and Karplus, M (1999). "Native proteins are surface-molten solids: application of the Lindemann criterion for the solid versus liquid state". J Mol Biol 285 (4): 1371–1375. doi:10.1006/jmbi.1998.2374. PMID 9917381.
- ^ Levitt M, Chothia C. (1976). "Structural patterns in globular proteins". Nature 261 (5561): 552–8. Bibcode 1976Natur.261..552L. doi:10.1038/261552a0. PMID 934293.
- ^ Hutchinson EG, Thornton JM. (1993). "The Greek key motif: extraction, classification and analysis". Protein Eng 6 (3): 233–45. doi:10.1093/protein/6.3.233. PMID 8506258.
- ^ Savageau MA. (1986). "Proteins of Escherichia coli come in sizes that are multiples of 14 kDa: domain concepts and evolutionary implications". Proc Natl Acad Sci USA 83 (5): 1198–202. doi:10.1073/pnas.83.5.1198. PMC 323042. PMID 3513170. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=3513170.
- ^ a b c Islam SA, Luo J, Sternberg MJ. (1995). "Identification and analysis of domains in proteins". Protein Eng 8 (6): 513–25. doi:10.1093/protein/8.6.513. PMID 8532675.
- ^ Wheelan, S. J. and Marchler-Bauer, A. and Bryant, S. H. (2000). "Domain size distributions can predict domain boundaries". Bioinformatics 16 (7): 613–618. doi:10.1093/bioinformatics/16.7.613. PMID 11038331.
- ^ a b Garel, J. (1992). "Folding of large proteins: Multidomain and multisubunit proteins". In Creighton, T.. Protein Folding (First ed.). New York: W.H. Freeman and Company. pp. 405–454. ISBN 071677027X.
- ^ Bennett MJ, Schlunegger MP, Eisenberg D. (1995). "3D domain swapping: a mechanism for oligomer assembly". Protein Sci 4 (12): 2455–68. doi:10.1002/pro.5560041202. PMC 2143041. PMID 8580836. http://www.proteinscience.org/cgi/pmidlookup?view=long&pmid=8580836.
- ^ Heringa J, Taylor WR. (1997). "Three-dimensional domain duplication, swapping and stealing". Curr Opin Struct Biol 7 (3): 416–21. doi:10.1016/S0959-440X(97)80060-7. PMID 9204285.
- ^ Jacob F. (1977). "Evolution and tinkering". Science 196 (4295): 1161–6. doi:10.1126/science.860134. PMID 860134.
- ^ Campbell ID, Downing AK. (1994). "Building protein structure and function from modular units". Trends Biotechnol 12 (5): 168–72. doi:10.1016/0167-7799(94)90078-7. PMID 7764899.
- ^ http://www.pdb.org/
- ^ Orengo CA, Jones DT, Thornton JM. (1994). "Protein superfamilies and domain superfolds". Nature 372 (6507): 631–4. doi:10.1038/372631a0. PMID 7990952.
- ^ Apic, G., Gough, J., and Teichmann, S. A (2001). "Domain combinations in archaeal, eubacterial and eukaryotic proteomes". J Mol Biol 310 (2): 311–325. doi:10.1006/jmbi.2001.4776. PMID 11428892.
- ^ Davidson JN, Chen KC, Jamison RS, Musmanno LA, Kern CB. (1993). "The evolutionary history of the first three enzymes in pyrimidine biosynthesis". Bioessays 15 (3): 157–64. doi:10.1002/bies.950150303. PMID 8098212.
- ^ Henikoff S, Greene EA, Pietrokovski S, Bork P, Attwood TK, Hood L. (1997). "Gene families: the taxonomy of protein paralogs and chimeras". Science 278 (5338): 609–14. doi:10.1126/science.278.5338.609. PMID 9381171.
- ^ Bork, P. and Doolittle, R. F (1992). "Proposed acquisition of an animal protein domain by bacteria2". Proc Natl Acad Sci USA 89 (19): 8990–8994. doi:10.1073/pnas.89.19.8990. PMC 50050. PMID 1409594. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=50050.
- ^ Heringa J. (1998). "Detection of internal repeats: how common are they?". Curr Opin Struct Biol 8 (3): 338–45. doi:10.1016/S0959-440X(98)80068-7. PMID 9666330.
- ^ Politou, A. S., Gautel, M., Improta, S., Vangelista, L., and Pastore, A (1996). "The elastic I-band region of titin is assembled in a 'modular' fashion by weakly interacting Ig-like domains". J Mol Biol 255 (4): 604–616. doi:10.1006/jmbi.1996.0050. PMID 8568900.
- ^ a b McLachlan, A. D (1979). "Gene duplications in the structural evolution of chymotrypsin". J Mol Biol 128 (1): 49–79. doi:10.1016/0022-2836(79)90308-5. PMID 430571.
- ^ Moore JD, Endow SA. (1996). "Kinesin proteins: a phylum of motors for microtubule-based motility". Bioessays 18 (3): 207–19. doi:10.1002/bies.950180308. PMID 8867735.
- ^ Russell, R. B (1994). "Domain insertion". Protein Eng 7 (12): 1407–1410. doi:10.1093/protein/7.12.1407. PMID 7716150.
- ^ Levinthal, C. (1968). "Are there pathways for protein folding?". J Chim Phys 65: 44–45. http://www.biochem.wisc.edu/courses/biochem704/Reading/Levinthal1968.pdf.
- ^ Dill KA. (1999). "Polymer principles and protein folding". Protein Sci 8 (6): 1166–80. doi:10.1110/ps.8.6.1166. PMC 2144345. PMID 10386867. http://www.proteinscience.org/cgi/pmidlookup?view=long&pmid=10386867.
- ^ Leopold PE, Montal M, Onuchic JN. (1992). "Protein folding funnels: a kinetic approach to the sequence-structure relationship". Proc Natl Acad Sci USA 89 (18): 8721–5. doi:10.1073/pnas.89.18.8721. PMC 49992. PMID 1528885. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=1528885.
- ^ Dill KA, Chan HS. (1997). "From Levinthal to pathways to funnels". Nat Struct Biol 4 (1): 10–9. doi:10.1038/nsb0197-10. PMID 8989315.
- ^ White SH, Jacobs RE. (1990). "Statistical distribution of hydrophobic residues along the length of protein chains. Implications for protein folding and evolution". Biophys J 57 (4): 911–21. Bibcode 1990BpJ....57..911W. doi:10.1016/S0006-3495(90)82611-4. PMC 1280792. PMID 2188687. http://www.biophysj.org/cgi/pmidlookup?view=long&pmid=2188687.
- ^ George RA, Heringa J. (2002). "SnapDRAGON: a method to delineate protein structural domains from sequence data". J Mol Biol 316 (3): 839–51. doi:10.1006/jmbi.2001.5387. PMID 11866536.
- ^ George RA, Lin K, Heringa J. (2005). "Scooby-domain: prediction of globular domains in protein sequence". Nucleic Acids Res 33 (Web Server issue): W160–3. doi:10.1093/nar/gki381. PMC 1160142. PMID 15980446. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1160142.
- ^ Desmadril, M. and Yon, J. M (1981). "Existence of intermediates in the refolding of T4 lysozyme at pH 7.4". Biochem Biophys Res Commun 101 (2): 563–569. doi:10.1016/0006-291X(81)91296-1. PMID 7306096.
- ^ Teale JM, Benjamin DC. (1977). "Antibody as immunological probe for studying refolding of bovine serum albumin. Refolding within each domain". J Biol Chem 252 (13): 4521–6. PMID 873903.
- ^ Creighton, T. E. (1983). Proteins: Structures and molecular properties. Freeman, New York. Second edition.
- ^ Bu Z, Callaway DJ (2011). "Proteins MOVE! Protein dynamics and long-range allostery in cell signaling". Advances in Protein Chemistry and Structural Biology 83: 163–221. doi:10.1016/B978-0-12-381262-9.00005-7. PMID 21570668. http://linkinghub.elsevier.com/retrieve/pii/B978-0-12-381262-9.00005-7.
- ^ Farago B, Li J, Cornilescu G, Callaway DJ, Bu Z (November 2010). "Activation of Nanoscale Allosteric Protein Domain Motion Revealed by Neutron Spin Echo Spectroscopy". Biophysical Journal 99 (10): 3473–3482. Bibcode 2010BpJ....99.3473F. doi:10.1016/j.bpj.2010.09.058. PMC 2980739. PMID 21081097. http://linkinghub.elsevier.com/retrieve/pii/S0006-3495(10)01208-7.
- ^ Bu Z, Biehl R, Monkenbusch M, Richter D, D.J.E. Callaway (2005). "Coupled protein domain motion in Taq polymerase revealed by neutron spin-echo spectroscopy.". Proc Natl Acad Sci USA 102 (49): 17646–17651. Bibcode 2005PNAS..10217646B. doi:10.1073/pnas.0503388102. PMC 1345721. PMID 16306270. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1345721.
- ^ a b c d Potestio, R., Pontiggia, F. and Micheletti, C. (2009). "Coarse-grained description of protein internal dynamics: an optimal strategy for decomposing proteins in rigid subunits.". Biophysical Journal 96 (12): 4993–5002. doi:10.1016/j.bpj.2009.03.051. PMC 2712024. PMID 19527659. http://www.cell.com/biophysj/abstract/S0006-3495%2809%2900781-4.
- ^ a b Gerstein M, Lesk AM, Chothia C. (1994). "Structural mechanisms for domain movements in proteins". Biochemistry 33 (22): 6739–49. doi:10.1021/bi00188a001. PMID 8204609.
- ^ Herzberg O et al. (1996). "Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites". Proc Natl Acad Sci USA 93 (7): 2652–7. doi:10.1073/pnas.93.7.2652. PMC 39685. PMID 8610096. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=8610096.
- ^ a b Janin, J. and Wodak, S. J (1983). "Structural domains in proteins and their role in the dynamics of protein function". Prog Biophys Mol Biol 42 (1): 21–78. doi:10.1016/0079-6107(83)90003-2. PMID 6353481.
- ^ a b Hayward, 1999
- ^ Meador, W. E., Means, A. R., and Quiocho, F. A (1992). "Target enzyme recognition by calmodulin: 2.4A structure of a calmodulin-peptide complex". Science 257 (5074): 1251–1255. doi:10.1126/science.1519061. PMID 1519061.
- ^ Ikura, M., Clore, G. M., Gronenborn, A. M., Zhu, G., Klee, C. B., and Bax, A (1992). "Solution structure of a calmodulin-target peptide complex by multidimensional NMR". Science 256 (5057): 632–638. doi:10.1126/science.1585175. PMID 1585175.
- ^ a b Sowdhamini R, Blundell TL. (1995). "An automatic method involving cluster analysis of secondary structures for the identification of domains in proteins". Protein Sci 4 (3): 506–20. doi:10.1002/pro.5560040317. PMC 2143076. PMID 7795532. http://www.proteinscience.org/cgi/pmidlookup?view=long&pmid=7795532.
- ^ Swindells, M. B (1995). "A procedure for detecting structural domains in proteins". Protein Sci 4 (1): 103–112. doi:10.1002/pro.5560040113. PMC 2142966. PMID 7773168. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2142966.
- ^ Tsai CJ, Nussinov R. (1997). "Hydrophobic folding units derived from dissimilar monomer structures and their interactions". Protein Sci 6 (1): 24–42. doi:10.1002/pro.5560060104. PMC 2143523. PMID 9007974. http://www.proteinscience.org/cgi/pmidlookup?view=long&pmid=9007974.
- ^ a b Crippen, G. M (1978). "The tree structural organisation of proteins". J Mol Biol 126 (3): 315–332. doi:10.1016/0022-2836(78)90043-8. PMID 745231.
- ^ Rossmann MG, Moras D, Olsen KW. (1974). "Chemical and biological evolution of nucleotide-binding protein". Nature 250 (463): 194–9. doi:10.1038/250194a0. PMID 4368490.
- ^ Rose GD. (1979). "Hierarchic organization of domains in globular proteins". J Mol Biol 134 (3): 447–70. doi:10.1016/0022-2836(79)90363-2. PMID 537072.
- ^ a b Go N, Taketomi H. (1978). "Respective roles of short- and long-range interactions in protein folding". Proc Natl Acad Sci USA 75 (2): 559–63. doi:10.1073/pnas.75.2.559. PMC 411294. PMID 273218. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=411294.
- ^ a b Holm L, Sander C. (1997). "Dali/FSSP classification of three-dimensional protein folds". Nucleic Acids Res 25 (1): 231–4. doi:10.1093/nar/25.1.231. PMC 146389. PMID 9016542. http://nar.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=9016542.
- ^ a b Siddiqui AS, Barton GJ. (1995). "Continuous and discontinuous domains: an algorithm for the automatic generation of reliable protein domain definitions". Protein Sci 4 (5): 872–84. doi:10.1002/pro.5560040507. PMC 2143117. PMID 7663343. http://www.proteinscience.org/cgi/pmidlookup?view=long&pmid=7663343.
- ^ Zehfus, M. H (1997). "Identification of compact, hydrophobically stabilized domains and modules containing multiple peptide chains". Protein Sci 6 (6): 1210–1219. doi:10.1002/pro.5560060609. PMC 2143719. PMID 9194181. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2143719.
- ^ Taylor WR. (1999). "Protein structural domain identification". Protein Eng 12 (3): 203–16. doi:10.1093/protein/12.3.203. PMID 10235621.
- ^ Wodak, S. J. and Janin, J (1981). "Location of structural domains in protein". Biochemistry 20 (23): 6544–6552. doi:10.1021/bi00526a005. PMID 7306523.
- ^ Rashin, 1985
- ^ Zehfus MH, Rose GD. (1986). "Compact units in proteins". Biochemistry 25 (19): 5759–65. doi:10.1021/bi00367a062. PMID 3778881.
- ^ a b Aleksiev, T., Potestio, R., Pontiggia, F., Cozzini, S. and Micheletti, C. (2009). "PiSQRD: a web server for decomposing proteins into quasi-rigid dynamical domains.". Bioinformatics 25 (20): 2743–4. doi:10.1093/bioinformatics/btp512. PMID 19696046. http://bioinformatics.oxfordjournals.org/cgi/content/abstract/btp512.
- ^ Micheletti, C., Carloni, P. and Maritan, A. Accurate and efficient description of protein vibrational dynamics: comparing molecular dynamics and gaussian models, Proteins, 55, 635, 2004.
- ^ Barclay A (2003). "Membrane proteins with immunoglobulin-like domains--a master superfamily of interaction molecules". Semin Immunol 15 (4): 215–23. doi:10.1016/S1044-5323(03)00047-2. PMID 14690046.
Key papers
- Berman HM et al. (2000). "The Protein Data Bank". Nucleic Acids Res 28 (1): 235–42. doi:10.1093/nar/28.1.235. PMC 102472. PMID 10592235. http://nar.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=10592235.
- Tooze, John; Brändén, Carl-Ivar (1999). Introduction to protein structure. New York: Garland Pub. ISBN 0-8153-2305-0.
- Chothia C. (1992). "Proteins. One thousand families for the molecular biologist". Nature 357 (6379): 543–4. doi:10.1038/357543a0. PMID 1608464.
- Das S, Smith TF. (2000). "Identifying nature's protein Lego set". Adv Protein Chem 54: 159–83. doi:10.1016/S0065-3233(00)54006-6. PMID 10829228.
- Dietmann S, Park J, Notredame C, Heger A, Lappe M, Holm L. (2001). "A fully automatic evolutionary classification of protein folds: Dali Domain Dictionary version 3". Nucleic Acids Res 29 (1): 55–7. doi:10.1093/nar/29.1.55. PMC 29815. PMID 11125048. http://nar.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=11125048.
- Dyson HJ, Sayre JR, Merutka G, Shin HC, Lerner RA, Wright PE. (1992). "Folding of peptide fragments comprising the complete sequence of proteins. Models for initiation of protein folding. II. Plastocyanin". J Mol Biol 226 (3): 819–35. doi:10.1016/0022-2836(92)90634-V. PMID 1507228.
- Fersht AR. (1997). "Nucleation mechanisms in protein folding". Curr Opin Struct Biol 7 (1): 3–9. doi:10.1016/S0959-440X(97)80002-4. PMID 9032066.
- George DG, Hunt LT, Barker WC. (1996). "PIR-International Protein Sequence Database". Methods Enzymol 266: 41–59. doi:10.1016/S0076-6879(96)66005-4. PMID 8743676.
- Go M. (1981). "Correlation of DNA exonic regions with protein structural units in haemoglobin". Nature 291 (5810): 90–2. doi:10.1038/291090a0. PMID 7231530.
- Hadley, C and Jones, D.T. (1999). "A systematic comparison of protein structure classifications: SCOP, CATH and FSSP". Struct Fold Des 7 (9): 1099–112. doi:10.1016/S0969-2126(99)80177-4. PMID 10508779.
- Hayward S. (1999). "Structural principles governing domain motions in proteins". Proteins 36 (4): 425–35. doi:10.1002/(SICI)1097-0134(19990901)36:4<425::AID-PROT6>3.0.CO;2-S. PMID 10450084.
- Heringa J, Argos P. (1991). "Side-chain clusters in protein structures and their role in protein folding". J Mol Biol 220 (1): 151–71. doi:10.1016/0022-2836(91)90388-M. PMID 2067014.
- Honig B. (1999). "Protein folding: from the levinthal paradox to structure prediction". J Mol Biol 293 (2): 283–93. doi:10.1006/jmbi.1999.3006. PMID 10550209.
- Kim PS, Baldwin RL. (1990). "Intermediates in the folding reactions of small proteins". Annu Rev Biochem 59 (1): 631–60. doi:10.1146/annurev.bi.59.070190.003215. PMID 2197986.
- Murvai J, Vlahovicek K, Barta E, Cataletto B, Pongor S. (2000). "The SBASE protein domain library, release 7.0: a collection of annotated protein sequence segments". Nucleic Acids Res 28 (1): 260–2. doi:10.1093/nar/28.1.260. PMC 102474. PMID 10592241. http://nar.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=10592241.
- Murzin AG, Brenner SE, Hubbard T, Chothia C. (1995). "SCOP: a structural classification of proteins database for the investigation of sequences and structures". J Mol Biol 247 (4): 536–40. doi:10.1016/S0022-2836(05)80134-2. PMID 7723011.
- Janin J, Chothia C. (1985). "Domains in proteins: definitions, location, and structural principles". Methods Enzymol 115: 420–30. doi:10.1016/0076-6879(85)15030-5. PMID 4079796.
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. (2000). "SMART: a web-based tool for the study of genetically mobile domains". Nucleic Acids Res 28 (1): 231–4. doi:10.1093/nar/28.1.231. PMC 102444. PMID 10592234. http://nar.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=10592234.
- Siddiqui AS, Dengler U, Barton GJ. (2001). "3Dee: a database of protein structural domains". Bioinformatics 17 (2): 200–1. doi:10.1093/bioinformatics/17.2.200. PMID 11238081.
- Srinivasarao GY, Yeh LS, Marzec CR, Orcutt BC, Barker WC, Pfeiffer F. (1999). "Database of protein sequence alignments: PIR-ALN". Nucleic Acids Res 27 (1): 284–5. doi:10.1093/nar/27.1.284. PMC 148157. PMID 9847202. http://nar.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=9847202.
- Tatusov RL et al. (2001). "The COG database: new developments in phylogenetic classification of proteins from complete genomes". Nucleic Acids Res 29 (1): 22–8. doi:10.1093/nar/29.1.22. PMC 29819. PMID 11125040. http://nar.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=11125040.
- Taylor WR, Orengo CA. (1989). "Protein structure alignment". J Mol Biol 208 (1): 1–22. doi:10.1016/0022-2836(89)90084-3. PMID 2769748.
- Yang AS, Honig B. (1995). "Free energy determinants of secondary structure formation: I. alpha-Helices". J Mol Biol 252 (3): 351–65. doi:10.1006/jmbi.1995.0502. PMID 7563056.
- Yang AS, Honig B. (1995). "Free energy determinants of secondary structure formation: II. Antiparallel beta-sheets". J Mol Biol 252 (3): 366–76. doi:10.1006/jmbi.1995.0503. PMID 7563057.
External links
- The Protein Families (Pfam) database clan browser provides easy access to information about protein structural domains. A clan contains two or more Pfam families that have arisen from a single evolutionary origin.
Structural domain databases
- Conserved Domains at the National Center for Biotechnology website
- 3Dee
- CATH
- DALI
- SCOP
- Pawson Lab - Protein interaction domains
- Nash Lab - Protein interaction domains in Signal Transduction
- Definition and assignment of structural domains in proteins.
Sequence domain databases
- InterPro
- Pfam
- PROSITE
- ProDom
- SMART
- NCBI Conserved Domain Database
- SUPERFAMILY Library of HMMs representing superfamilies and database of (superfamily and family) annotations for all completely sequenced organisms
Protein domains General All-α folds: All-β folds: α/β folds: α+β folds: Irregular folds: ←Secondary structureProcesses Structures Types List of types of proteins · List of proteins · Membrane protein · Globular protein (Globulin, Albumin) · Fibrous proteinbiochemical families: prot · nucl · carb (glpr, alco, glys) · lipd (fata/i, phld, strd, gllp, eico) · amac/i · ncbs/i · ttpy/i
B proteins: BY STRUCTURE: membrane, globular (en, ca, an), fibrousProtein structure Nucleic acid structure See also Protein · Protein domain · Protein engineering · Nucleic acid · DNA · RNA · Nucleic acid double helixCategories:- Protein structure
- Protein domains
Wikimedia Foundation. 2010.